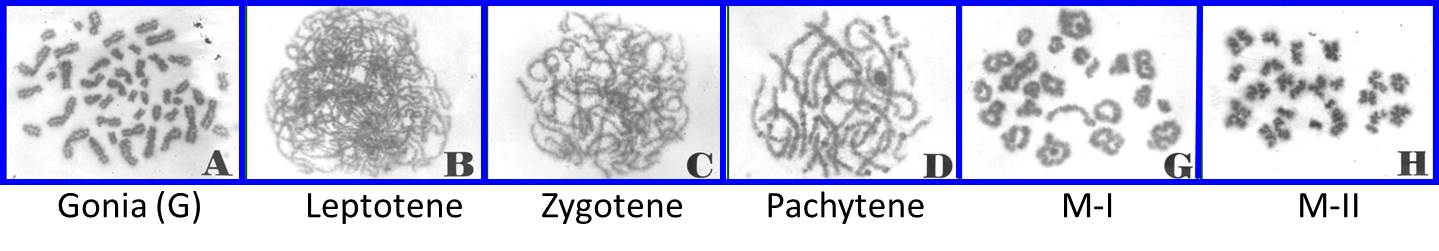
| [1] Suppression of meiotic progression by de novo translocation (Commentary) |
| Meiotic progression in human testis |
 |
Balanced reciprocal translocations, recTr, are the most common structural
chromosome abnormalities found in the general populations. The reciprocal
translocations themselves have been thought to be genetically inactive
but could result in the outcome of miscarriage or inborn abnormalities
when its meiotic segregants (products of its adjacent segregation or 3:1
segregation) are inherited by offspring. In this respect, the induction
of translocations in germ cells called attention to the hereditary effects
of radiation. The induction of translocations by radiation in spermatogonial
stem cells has been extensively studied in the experimental animals (Translocations
in spermatogonia, mousae and other mammals). In spite of the experimental evidence accumulated in various mammalian
species including man, the inheritance of translocations has not been confirmed
in the offspring of Atomic bomb survivors in Hiroshima and Nagasaki. The
enigma has not been solved.
Recently, evidence has been accumulated to show that large fraction of de novo translocations, newly formed but
not inherited, translocations are accumulated in the infertile men who are
diagnosed to suffer from azoosperma and oligospermia (Infertile men, and also recent reviews by Mau-Holzmann, 2005; Madan, 2012; Godo et al., 2013; Zhang et al., 2015).
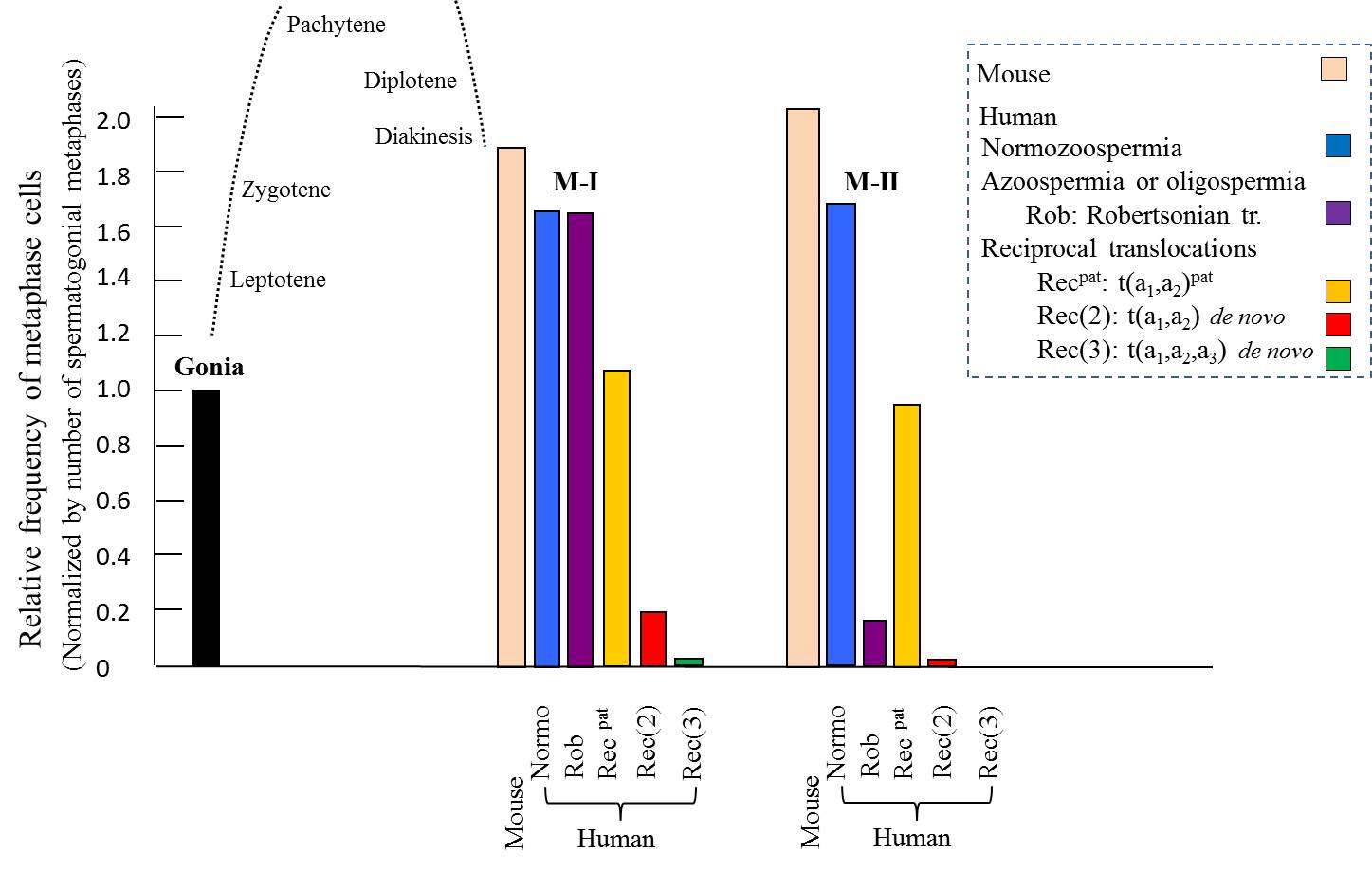 |
The
above figure is a summary of the meiotic progression in 8 infertile men with azoospermia
or oligospermia. In the carriers of de
novo recTr, the meiotic progression from spermatogonia/pachytene to
metaphase I, M-I, is severely suppressed and the suppresstion is further
evident for stage from M-I to M-II. However, when the translocation is
inherited from his parent, the suppressive effects are minimal. For the
Robertsonian translocations of de novo
origin, progress towards M-I is not suppressed but progression from M-I to M-II
is suppressed.
The suppression of meiotic progression by reciprocal translocation of de novo origin has been implicate for the humped dose-response curves of radiation-induced
spermatogonial translocations, and reduced transmission of radiation-induced
translocations to the offspring of A-bomb survivors (Sasaki, 2006).
|
References cited |
|
Mau-Holzmann, U. A. (2005): Somatic chromosomal
abnormalities in infertile men and women. Cytogenet. Genome Res., 111:317-336. |
| [2] Sex chromosome aneuploidy in offspring of A-bomb survivors (Commentary) |
In the F1 children bone to the exposed parent, non-Robertsonian structural aberrations are found in 0.16 % of the exposed parents, which is comparable to that in the newborn infants (0.14 % of world total and 0.10 % in Japanese children) in general populations and lower than that in the internal controls (unexposed, 0.26%). Since parental origin of these chromosome aberrations has not been fully determined, the contribution of the radiation dose cannot be assessed with any certainty (see, A-bomb F1 study).
On the other hand, the sex chromosome
aneuploidies are all de novo mutations. However, in the experimental animals including Drosophila,
aneuploidy has been generally accepted to have no relation to the radiation
dose. Indeed, the frequencies of sex chromosome aneuploidy are very comparable
among newborns in general populations, children bone to exposed and unexposed
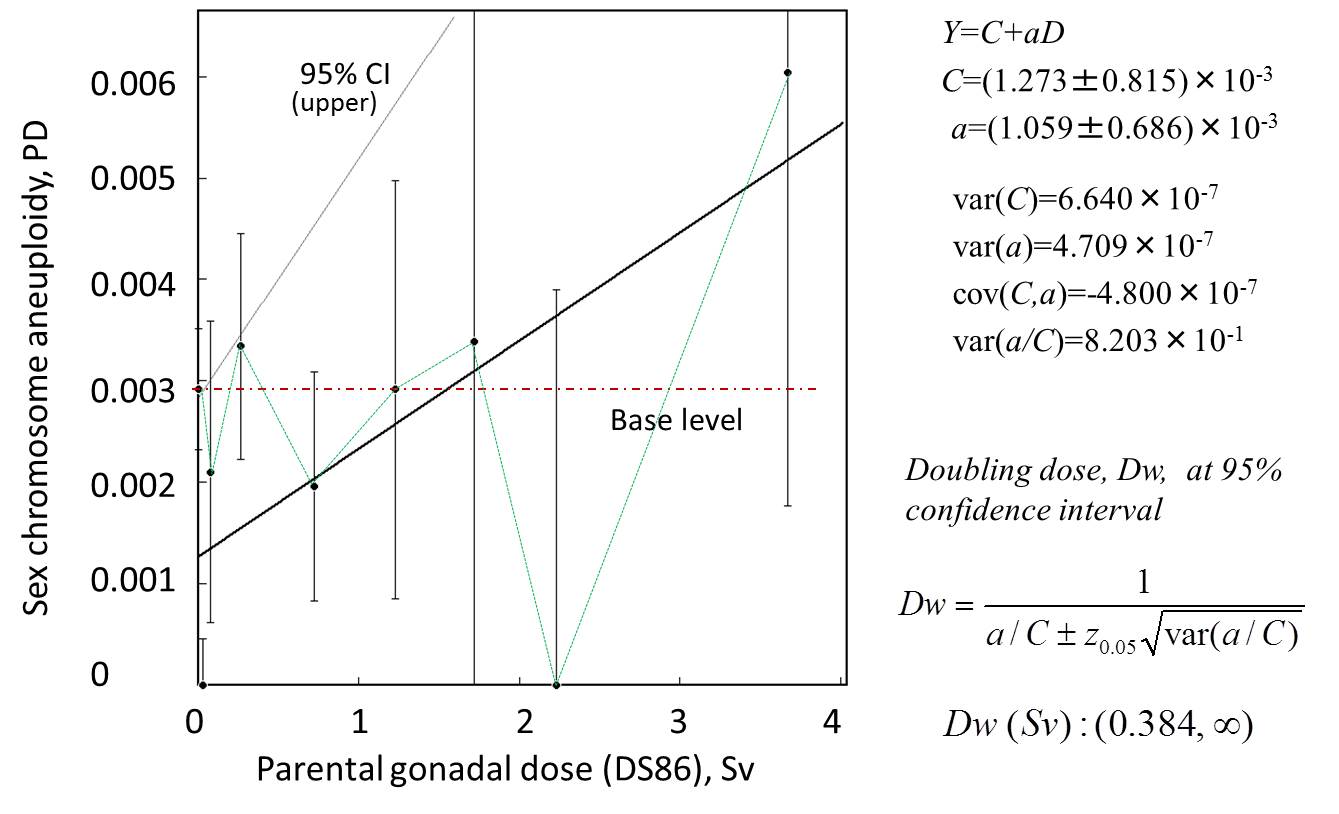
parents. Simple dose response for absolute frequencies shows the dose-response
with minimum doubling dose of about Dw=0.28 Sv. However, the relative risk is dose
independent, that is Dw=∞. Another point is that the aneuploidy is also dependent on the maternal
age. Thus, the doubling dose should be treated with caution. With these
modifying factors in considerations, Neel et al. (1990) concluded that
the exposuer did not produce and increase in aneuplidy in this population.
Neel, J. V., Schull, W. J., Awa, A. A., Satoh, C., Kato, H., Otake, M. and Yoshimoto, Y. (1990): The children of parents exposed to atomic bombs: estimates of the genetic doubling dose of radiation for humans. Am. J. Hum. Genet., 46:1053-1072. |
 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||